An introduction to statistics in R
A series of tutorials by Mark Peterson for working in R
Chapter Navigation
- Basics of Data in R
- Plotting and evaluating one categorical variable
- Plotting and evaluating two categorical variables
- Analyzing shape and center of one quantitative variable
- Analyzing the spread of one quantitative variable
- Relationships between quantitative and categorical data
- Relationships between two quantitative variables
- Final Thoughts on linear regression
- A bit off topic - functions, grep, and colors
- Sampling and Loops
- Confidence Intervals
- Bootstrapping
- More on Bootstrapping
- Hypothesis testing and p-values
- Differences in proportions and statistical thresholds
- Hypothesis testing for means
- Final thoughts on hypothesis testing
- Approximating with a distribution model
- Using the normal model in practice
- Approximating for a single proportion
- Null distribution for a single proportion and limitations
- Approximating for a single mean
- CI and hypothesis tests for a single mean
- Approximating a difference in proportions
- Hypothesis test for a difference in proportions
- Difference in means
- Difference in means - Hypothesis testing and paired differences
- Shortcuts
- Testing categorical variables with Chi-sqare
- Testing proportions in groups
- Comparing the means of many groups
- Linear Regression
- Multiple Regression
- Basic Probability
- Random variables
- Conditional Probability
- Bayesian Analysis
Basics of Data in R
1.1 Background
When starting out with statistics and data analysis, you may have calculated many statistics by hand, possibly with a calculator or spreadsheet. By-hand calculations are a great starting point to understand basic concepts and acquaint yourself with data analysis. However, as data sets grow larger and statistical questions grow more complex, being able to run your analyses in a computer program will become increasingly important. Here, we are going to explore one of these tools: R, a statistics environment designed to be a fully operational programming language.
1.1.1 Why R?
R was designed with high quality graphics explicitly in mind, which makes the default plots it produces elegant while allowing users full control over the outputs. In addition, this flexibility extends to the handling and display of raw data, making it possible to store, manipulate, and analyze a wide variety of data types in a single program. This flexibility comes at the cost of a large learning curve, especially as many of you are probably new to computer programming. Finally, R is free, both free as in beer (no cost) and free as in speech (the source code is available, can be manipulated, and can run on any platform), making it an accessible choice for students and researchers.
RStudio provides a useful wrapper for R – it allows us to save what we are doing each step of the way, and displays useful information (graphs, help files, etc.) alongside our progress. For all of our activities, we will be working in RStudio.
R can be daunting when you first start, but it is no more difficult to learn or use than any other new piece of software. The early tasks you want to accomplish in R are usually very easy … once you know what you are trying to do. The challenge of learning R is in changing your thinking and approach. By building your skills slowly, and making the process of using R explicit through this tutorial approach, the learning curve will be easy – even if it doesn’t initially seem that way.
1.1.2 Chapter Goals
- How to install and utilize a statistical package
- Be able to import data into R
- How to comfortably approach command line and computer code problems
1.2 Installing R and RStudio
Below are basic instructions for installing R and RStudio on each Linux, Mac, and even Windows. However, you can vist http://cran.r-project.org/bin for more details on the installation of R and http://www.rstudio.com/ide/download/desktop for information on RStudio.
1.2.1 Install in Linux
To install R on a Linux distribution, there are two main approaches. You can either download precompiled binaries for your specific installation, or compile yourself from source code. Binaries (which I recommend for most users) and information on how to install them are available at: http://lib.stat.cmu.edu/R/CRAN/bin/linux/.
RStudio binaries are available for Fedora and Ubuntu/Debian. Copy the current link for your system (examples below) from http://www.rstudio.com/ide/download/desktop, and install with either of the below (for Fedora 32-bit or Debian 64-bit respectively):
yum install http://download1.rstudio.org/rstudio-0.98.1091-i686.rpm
apt-get install http://download1.rstudio.org/rstudio-0.98.1091-amd64.debNote that these versions are likely out of date by the time you are reading this, so make sure to check the website for the most current versions and your platform. You will also likely need root privileges, so will likely need to run the above with sudo or as super-user.
Alternatively, a tarball of the source code is available for both R and RStudio at their respective download pages, if you prefer to install from source.
1.2.2 Install in Mac
In Mac, download the current version of R from http://cran.r-project.org/bin/macosx/ (currently R-3.1.2). Make sure to get the package for your particular OS version. This can be installed like other .pkg files – double-click and follow the on screen directions.
To install Rstudio on a Mac, download the latest .dmg RStudio file (currently 0.98.1091) from http://www.rstudio.com/ide/download/desktop. Install by mounting the .dmg file (double-click on it). An app should appear in a new Finder window. Drag it from the new window into your “Applications” folder. From now on, you can open RStudio directly from your Applications.
1.2.3 Install in Windows
In Windows, download the current version of R (currently R-3.1.2.exe) from http://cran.r-project.org/bin/windows/base/ and install like other .exe files: double-click and follow the on screen directions.
RStudio for Windows can be installed by downloading the latest .exe file (currently RStudio 0.98.1091) from http://www.rstudio.com/ide/download/desktop. Again, double-click and follow the on screen instructions.
1.3 Getting started
First, open RStudio as you would any other program you’ve installed. This is an integrated development environment (IDE) for R that will help keep all of your information together in one place. when you first open RStudio, it should look something like:
If your screen doesn’t look approximately like that, double check that you opened RStudio and not just R. You can work directly in R, but a number of the features we will use heavily will work much better in RStudio.
We are now all set to go and get started playing in R. Right off the bat, we are going to see one of the major differences between R and other computer programs you are used to (both for stats and all other programs). R is a sripting based language, which means that it is easy to keep track of everything we do, simply by saving a plain text file (like a .txt file) with our commands. This file will:
- Provide you with a record of everything you do in R
- Explicitly show all of your work
- Allow assessment of your progress
- Allow us to fix small mistakes in long pieces of code without doing everything
- Provide you access to previously used and learned commands
- Give you an easy way to turn in homework or quizzes or share data analysis
These last three points are especially powerful in R. Often times, you may find that there is a small mistake in your data after you have done a lot of work. In other programs, that might mean re-tracing all of your steps to re-create what you have done. In R, you just go back, fix the early mistake, and re-run what you already saved. This is even more powerful when, like in homework assignments or long-term data analysis projects, you need to do the same thing over again with completely different data. For all of these, it is then easy to share everything you did with an instructor, collaborator, or mentor to show all of the steps you have done.
1.3.1 Create a script file
To get started on your first R file, click on the File menu, then New and R Script. Next, we are going to save the file. Click on File then Save (or type Ctrl + s). In the dialog that pops up, navigate to the directory (folder) where you want to save your R work. This might be something like statsClass or learningR. Before you save it though, click on Create Folder and create a directory (folder) named scripts. Now, type a name for you script that includes your last name and ending with .R (e.g. “lastName_learningR.R”) and save it in your scripts directory. (This extra directory will come in handy later, when you have several scripts to keep organized.)
1.3.2 Notes to our future selves
Now, lets start by making some notes to ourselves. In computer programming, including R, these are called “comments” In R, anything that is preceded by a “#” is ignored. This lets us make comments, which can be incredibly helpful if we ever need to come back to the code, or share it with others. If you are doing this tutorial for a class, at the end of this chapter, you will each submit your script, along with an additional file we will produce near the end of the lab. These notes will help your instructor see what you did, and will help you when you need to use some of these commands again for the next chapter.
A note on convention: every command I tell you to type will look like the box below. In general, you can copy what you see below directly into your script file; however, things in ALL-CAPS should be replaced with your information.
# Script written by FIRSTNAME LASTNAME
# Learning R First Script
# YEAR-MON-DDThis information will help to remind you of what you were doing when writing the code, what its purpose is/was, and when it was written. Believe me, when you come back to this to work on your projects, you will appreciate having these comments. Comments can also help others to understand your code, and should become a habit for you. These directions will include comments throughout, but you should also add your own notes to the code as well. Part of the grade for this assignment, if you are doing this as part of a class, will include your comments and answers to questions in this document.
1.3.3 Running commands
Now, we need to see how these commands are actually passed to R. In RStudio, if you press Ctrl+Enter, it will send the highlighted text (or current line if nothing is highlighted) to be run by R. Highlight the three lines of comments (either with the mouse, or by holding shift and using the arrow keys) and press Ctrl+Enter. This action sends the comments to the Console and executes any code. Alternatively, you can click Run above the script window to accomplish the same thing. Here, the text is commented, so nothing happens.
1.4 Doing simple arithmetic in R
Before we go further, let’s just get a feel for what R does. First do something simple, type the following in your R script:
3 + 2You’ll noticed that nothing happened – that is because we haven’t told R to do anything with it yet. Now, put (or leave) your cursor (the vertical line that shows where you are typing) on that line and type Ctrl+Enter (hold Ctrl and type Enter). Now, the number 5 should have shown up, along with your command and some other “stuff” down in the “Console” Window.
We will discuss some of that other “stuff” in a bit. For now, what we see is that the “Console” window (in the lower-left corner by default in RStudio) is where R is actually “working.” The script file (in the upper-left, where we will do most of our typing) is just holding commands ready for R. (As an aside: you can work in R directly this way, but it makes it harder to track and repeat what you have done.)
Let’s try a little more arithmetic before we move on. First copy-paste your 3 + 2 line a few times. Then, replace the + with -, *, and / in separate lines. The copy-paste step saves you some typing. While it isn’t much here, getting into the habit early is a good idea. You can run all of these lines at once by highlighting all of them (either with your cursor or by holding Shift and hitting the arrow keys) then typing Ctrl+Enter. You should now see:
# Try some simple arithmetic
3 + 2## [1] 53 - 2## [1] 13 * 2## [1] 63 / 2## [1] 1.5Now that we have a feel for how to use R, let’s start talking about using variables – the way that R let’s us store information.
1.5 Entering Simple Data
The most important thing to be able to do in any stats program is to import your data. In R, there are several methods for doing so, and we will focus on just a few of them. Variables in R are very flexible and can take many different forms. Variable names must begin with a letter, but then can be any length and can include numerals. In R, an arrow, composed of a “less than” symbol and a dash (<-) is used to assign variables. Try it with this simple example:
# Store a simple variable, then display it
testVariable <- 3
testVariableDon’t forget to execute the command (with Ctrl+Enter) to see the results (which should just be the number 3). The first command creates a variable, and the last line tells the console to display it. This variable can now be used just like a number. Try the code below to see:
# Use the variable
testVariable + 2
testVariable * 2Note that the results are the same as if we had just used 3 instead of using testVariable. Now, to see one reason we are using an R script – go back and change the number you assigned to testVariable (that is, change the 3 in the line testVariable <- 3) to any other number you want. Then, re-run all of the lines after it. Now, instead of having to change the 3 everywhere that it occurred, you only need to change it once and can immediately see the changes. In real data analysis, this might be for a number of reasons, such as: you realized you made a typo, you collected more data, you are playing with different parameters for something, or you are running the same analysis on different sets of data.
1.6 Entering Longer Data
A single value is not usually very useful for statistics. You will often have multiple observations, and will want to keep them all together, rather than saving each as a different variable. R may have already given you a hint that it can do this. When you displayed the value of testVariable, you likely noticed a [1] next to the value. This is an “index” telling us that the value displayed is the first element of that variable. (For those with programming experience in other languages, note that the index starts at 1, not 0.) That is because, in R, most simple variables are stored as “vectors.” This allows us to store many values in a single variable. We need to tell R that we are giving it several values to store together using the function c(). Try it out with this code:
# Make a test vector, then display it
testVector <- c(1,3,4,6,8,10,15)
testVectorThis is our first time encountering a function in R. So, let’s take a moment to talk about them. Functions in R always work by calling the name of the function (in this case c which is short for “concatenate”) followed by a series of “arguments” (things for the function to use) separated by commas inside of parentheses. There are a lot of functions built-in to R (available immediately), and a nearly infinite number more available to be added (something we will cover soon). In addition, you can even write your own functions, something explored in a later chapter. For now, what you need to know is that a function takes an argument (or more than one) and does a series of commands on it, then returns an output. Here, c takes a series of numbers, puts them together, and returns them as a single vector.
Now, back to vectors. Vectors in R have some special properties, and these properties will become very useful. First, lets see how vectors handle a little bit of arithmetic:
# See how vectors respond to arithmetic
testVector + 3
testVector * 2
testVector + testVectorWhat did you notice about the outputs? For arguments that are only one number long, the same procedure is applied to each element (item) of testVector. When the arguments are the same length (have the same number of stored values) as testVector (e.g., itself), then the argument is applied element-for-element (as in the last line). This is very useful when we start comparing vectors against each other.
1.7 Playing with Data
Now that we have some sense about how these things work, we can begin to look at the data a little more closely. First, lets create a new vector with each element being 3 greater than the elements in testVector. Copy the first arithmetic line with testVector, change it to match the line below, and run it:
# Create a second vector
testVectorB <- testVector + 3
testVectorBAll outputs in R can be saved in this fashion, which allows you to go back and use them later or to perform other manipulations on them. For example, what if you only want to know what the 3rd element of testVectorB is? R allows us to do that using brackets [ ] to tell it which element we want to see. Try it:
# Display only the 3rd element of a vector
testVectorB[3]We can also use this to display only elements that meet certain criteria. Let’s display only the elements that are greater than 8:
# Display only elements greater than 8
testVectorB[testVectorB > 8]## [1] 9 11 13 18# Show how it is done:
testVectorB > 8## [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUEAs you can see from the last line, R produces a type of value called a “logical” that says whether or not a condition (here, being greater than 8) is met. When placed within brackets, only those elements that meet the condition are displayed. This is useful for displaying certain data, but is especially important when looking at two different variables. For example:
# Display the value of testVectorB when testVector equals 3
# Note that two '=' are used for logical tests
testVectorB[testVector == 3]As we begin to analyze more complex data (or data you have generated), we will have grouping variables (e.g., categories, such as a treatment) that can be used to split the data just like this.
Finally, to show the relationship between these two variables more clearly, we are going to generate a plot using these two vectors. The general format for plotting is to give the x-coordinates, followed by the y-coordinates, as follows:
# Make a plot of two vectors
plot(testVector, testVectorB)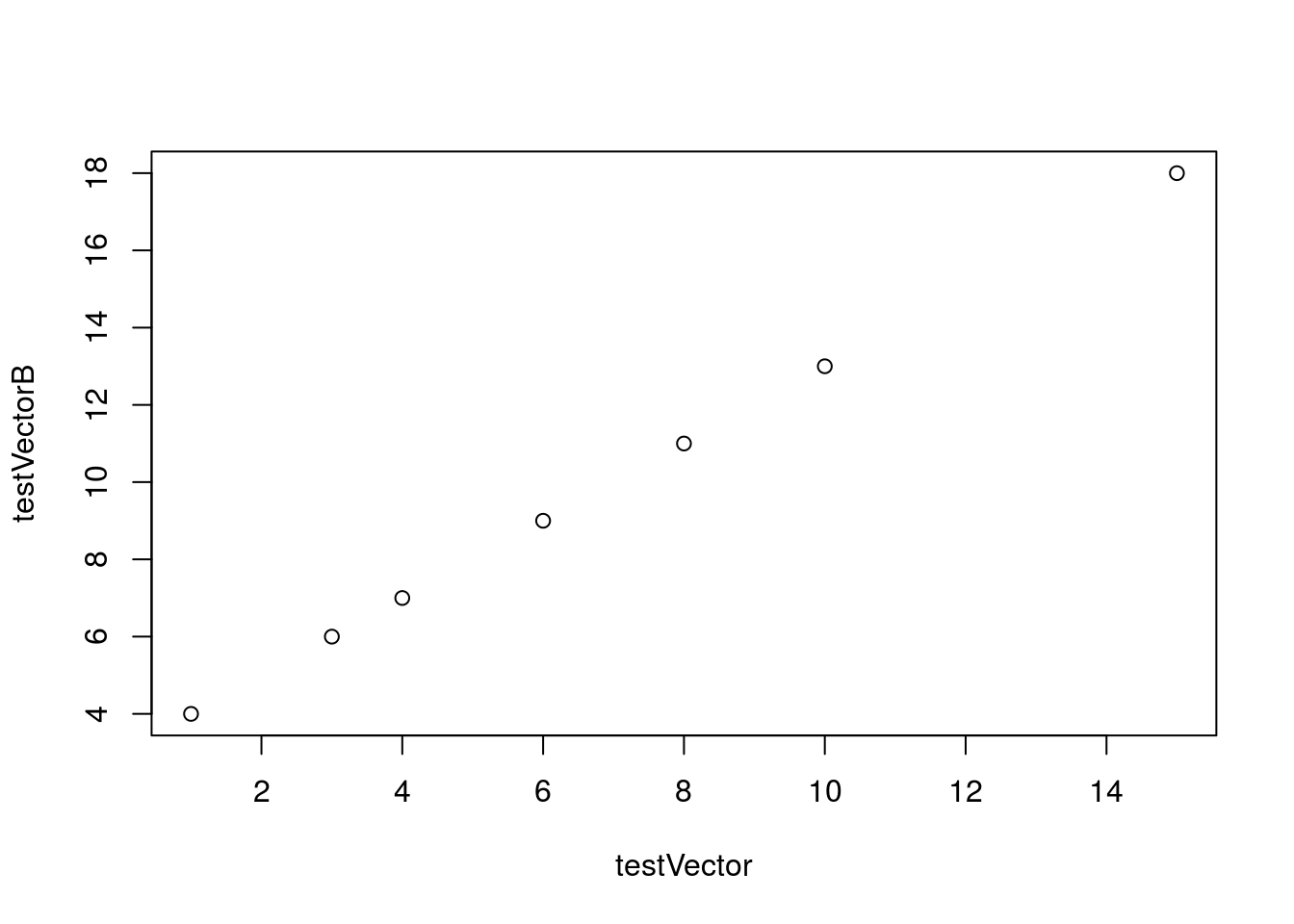
There are several additional options that we can use to change the plot, and we will explore those as we move forward with analyzing and plotting data.
1.8 Spreadsheet style data
Typing in data, as described above, works for simple datasets. What about when you have 100 observations, each with multiple measurements and properties? You could go through and type them all as individual vectors, but that would get cumbersome rapidly, and leads to typos and other errors affecting your results. Instead, we can use a type of variable called a data frame (data.frame in R) that will keep each vector together and reduce errors. Try this:
testDataFrame <- data.frame(testVector,testVectorB)
testDataFrameAs you can see, the rows now show which elements of each vector correspond with each other. In this way, you can see the relationship between the two vectors, and, if one were a grouping factor (e.g., treatment), use that relationship in your analyses. However, this still involved typing in all of your data, which is still a pain and error-prone. For any large dataset, you probably already have a spreadsheet (something you might open in Google Sheets, LibreOffice Calc, or Excel), and would like to be able to just use that. A safe, universally compatible approach, is to use a text based format for saving your data, such as comma-separated values (csv).
To use these files, create a directory (folder) named data in the same directory as your scripts directory. Then, click on the link(s) for the new data files as we encounter them and save them into your new data directory. For the first file, you should be able to download it from here: hotdogs.csv.
Another aside: A similar layout is generally great for any class or research projects. This keeps your data all together, and your analysis scripts nearby. I often also create an analysis directory in this same place to hold any outputs I generate. You can easily convert spreadsheet data to csv format for this as well, if your data aren’t already in plain-text format. Simply open your .ods, .xls, or similar file in your normal spreadsheet program, then select either “Save as” or “export” and look for an option to save as csv. Save the file in your data directory, and you should be readily able to import to R. (It is possible to open .xls files directly in R, but the proprietary nature of the format can often cause odd results.)
1.8.2 Import data directly
Often, especially when we are going to use many data sets (or the same data set in multiple scripts), it makes more sense to call the read.csv() function directly. Here, we will walk through a few tricks and tips that will make that easier.
The first thing to point out is that the line we copied above is really long, and probably different for each of you. If you have to type all that everytime, you may quickly mutiny and abandon R altogether. In addition, you may often want to share scripts. This may be because you are collaborating, reporting some finding to someone, or just turning in an assingment. In any of these cases, that long path (list of directories to get to the file) will be wrong. Instead, we will tell R where to look, then give it a much shorter path that is relative to that location. Then we only need to change that starting point.
First, we need to tell R where it should be looking for your documents (and where it should save outputs – we will deal with that later). This is similar to opening a folder, but we need to tell R to do it in text. This is called “setting the working directory”, and it will be the folder in which R opens and saves any files that you use, until you change the directory.
In RStudio, this can be done from a drop-down menu. Click Session,Set Working Directory, then To Source File Location. This will set the directory to the folder in which your script file (where you are typing commands) is located. If you want to select a different directory, click instead on Choose Directory (though that will make other commands I show you not work). You will notice that a command showed up in the terminal window, which should read something similar to:
# Manually set working directory
# Make sure that this is set for your computer, not mine
setwd('~/Documents/learningR/scripts')Copy this line (from your output, not the tutorial) into your script near the very top. Usually (and everytime after the next chapter) when you set the working directory, always put it near the very top of your script. That way, if you are working on a different computer (or sharing your script) it is easy to see (and change) the working directory to something that works for the local computer.
Now, we can load the file directly from text. Here is our first set of tricks. Type a comment saying you are loading the file directly, then type read.csv(" Notice that RStudio will automatically close the parenthesis and quote, but we still want to be typing inside of them. Next, type ../ (that is two periods followed by a slash). This is computer-speak for “look one directory (folder) above where I am”. We need to do this because we are in the scripts directory, and need to get one directory “over” to data.
Next, hit Tab (the tab key, not the letters). Notice that a menu pop-up menu opened with the names of things in that directory. Use the arrow keys to move to data, hit Enter, and R will fill in that part of the path. Finally, type ho and hit tab again. Select hotdogs.csv and hit Enter again (if it didn’t automatically complete all the way).
You should now have a line very similar to the one below. Exectute the line (with Ctrl+Enter) and see what happens.
# Read in the data directly
read.csv("../data/hotdogs.csv")You likely saw 54 rows of data scroll past, but it didn’t actually save the data. You need to tell R where to save it. Try the below:
# Read in the data directly
hotdogs <- read.csv("../data/hotdogs.csv")Now, you have saved the hotdog data as the variable hotdogs. You could, in principle, name this variable anything you would like. For your own analysis, that is fine. However, for these tutorials (and for sharing scripts) it helps to name them the same thing they are called in the book. Usually, this will mean naming the variable the same as the file name (with out the “.csv”), though occasionally we will use something different.
1.8.3 Looking at imported data
In the next chapter, we will start plotting data, including this data set. For now, we just want to look at the data to make sure that it read into R correctly.
The simplest function to do this is head(). This function shows us the first six rows (by default; other values are possible) of a data.frame. So, let’s try it on the hotdogs data:
# Look at the top of the hotdogs data
head(hotdogs)## Type Calories Sodium
## 1 Beef 186 495
## 2 Beef 181 477
## 3 Beef 176 425
## 4 Beef 149 322
## 5 Beef 184 482
## 6 Beef 190 587Here, we can see that we have 3 columns: Type, Calories, and Sodium. Beyond that, we don’t get much information, but at least we can see the names of each column and a bit of information about what type of data is in each.
To get a bit more information, we can use the function summary(). This function works on several types of variables in R (we will encounter it again soon). For data.frames, it gives us a bit of information about each column, as we can see:
# Look at the data a little more completely
summary(hotdogs)## Type Calories Sodium
## Beef :20 Min. : 86.0 Min. :144.0
## Meat :17 1st Qu.:132.0 1st Qu.:362.5
## Poultry:17 Median :145.0 Median :405.0
## Mean :145.4 Mean :424.8
## 3rd Qu.:172.8 3rd Qu.:503.5
## Max. :195.0 Max. :645.0Here, we see that there are three “Types” of hotdogs, that Calories range from 86 to 195, and that the mean amount of Sodium is 424.8. We will explore more about these things in the next chapter.
1.9 Generate the output for this tutorial.
For your assignment, you will create an html output from the script to confirm that your code worked. Even if you are not doing this tutorial for a class, generating this file will allow you to check your own work, and will catch most common mistakes. RStudio has a built in tool to generate these files, but it requires an R package called knitr. Install the knitr package by entering:
# Install knitr
# Don't forget to comment this out
install.packages('knitr')… and follow the on screen prompts.
After it is installed, you will want to put a # in front of this line . This is called “commenting out” and is a way of keeping R from trying to re-run a command (e.g., when you make this output, or if you run your script again) while still keeping a record of what you did.
Finally, click on the small notebook icon just above your script. It should say “Compile an HTML notebook from the current R script” when you mouse over it. Assign an appropriate title, and put your name as the author. Click Compile and an html file will be created. Submit these as instructed for the course. If you are in my class, all of your homework, quizzes, and exams will be submitted in this same way.